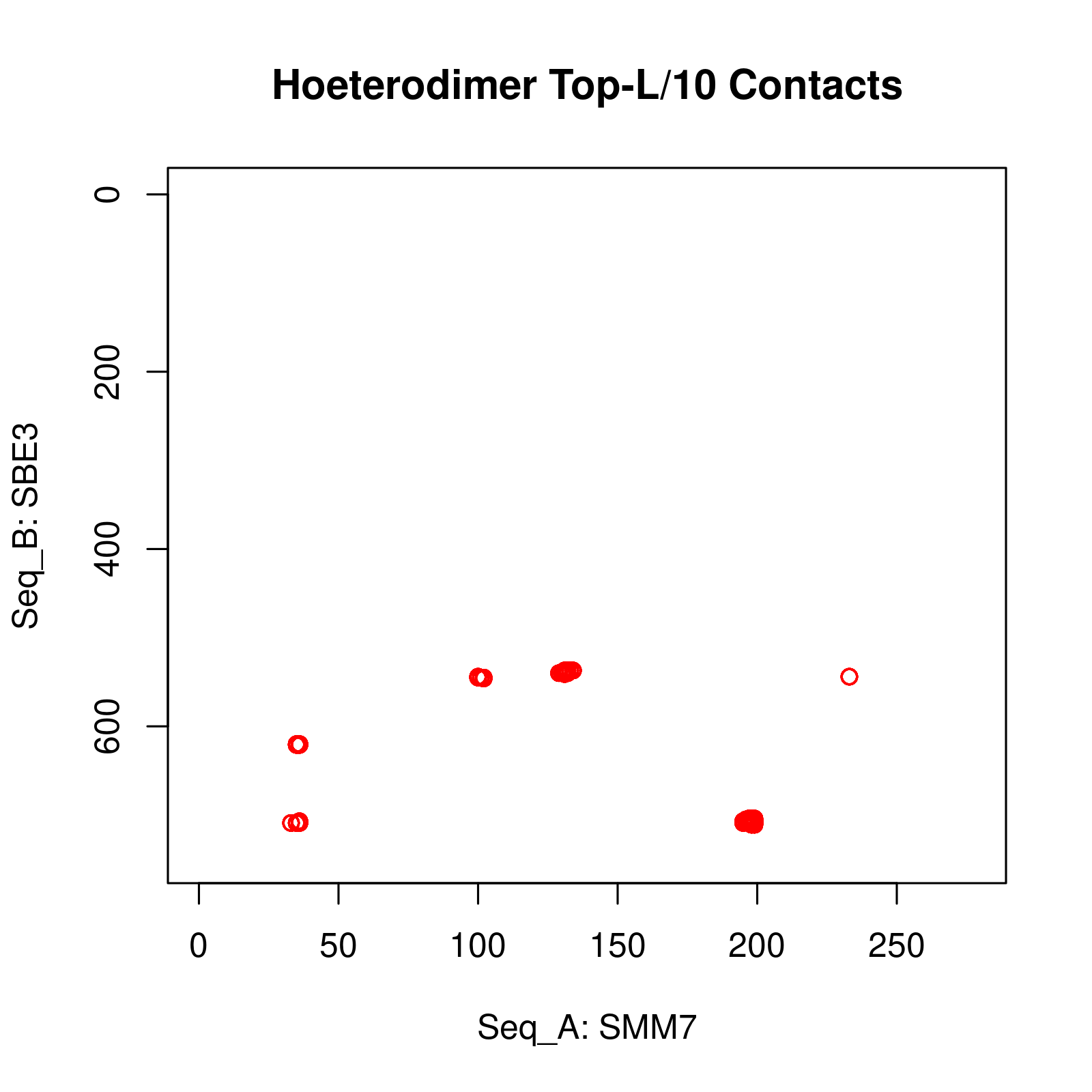
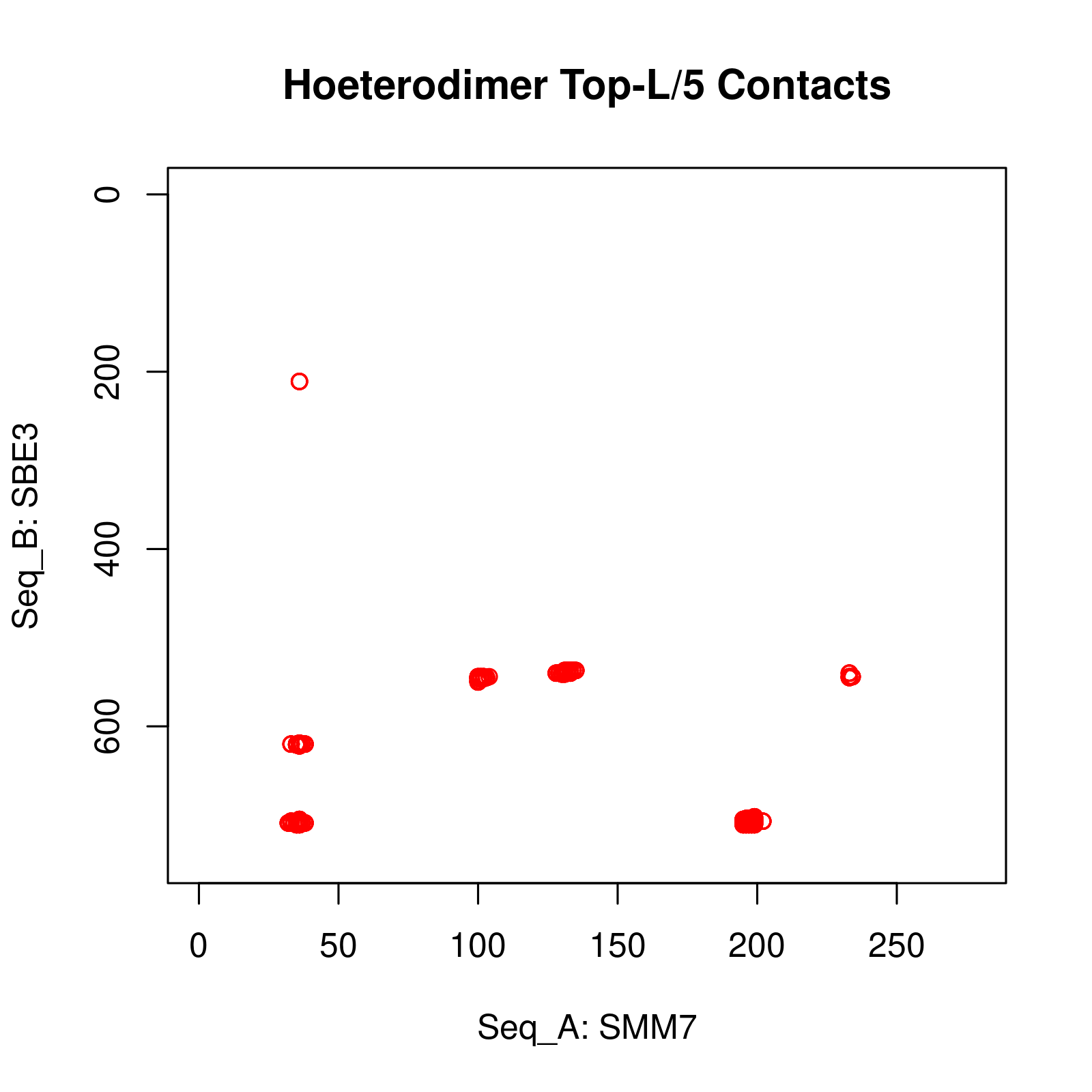
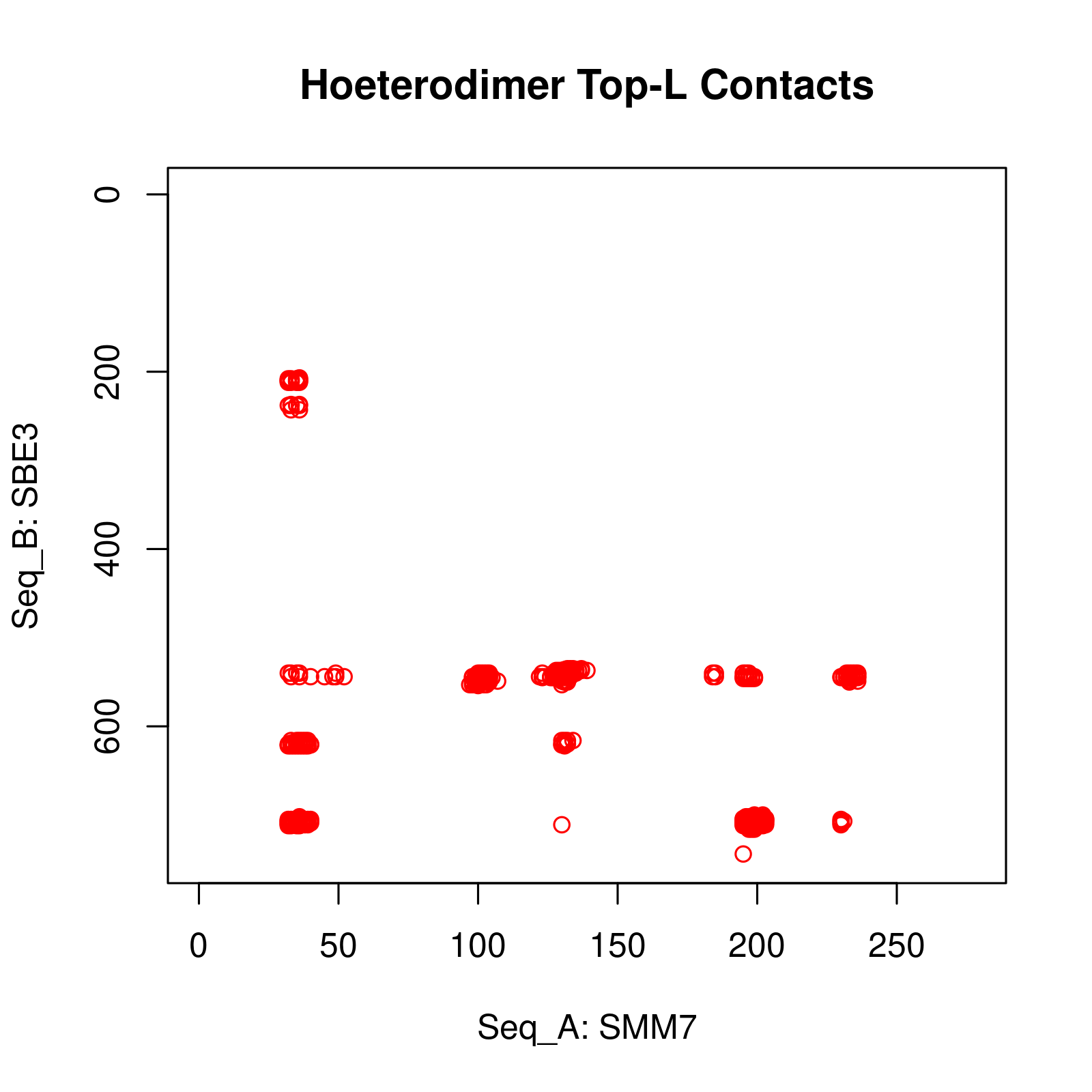
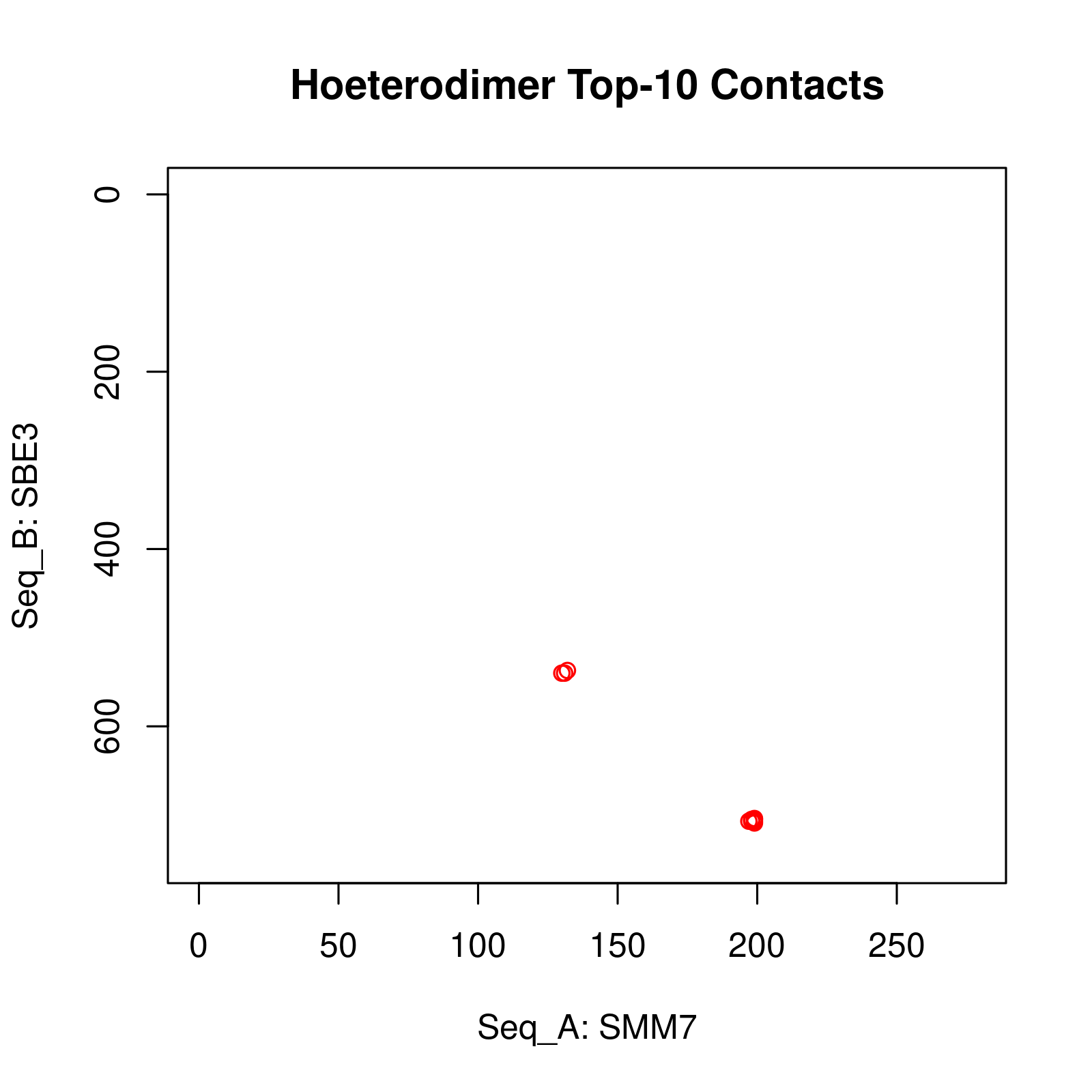
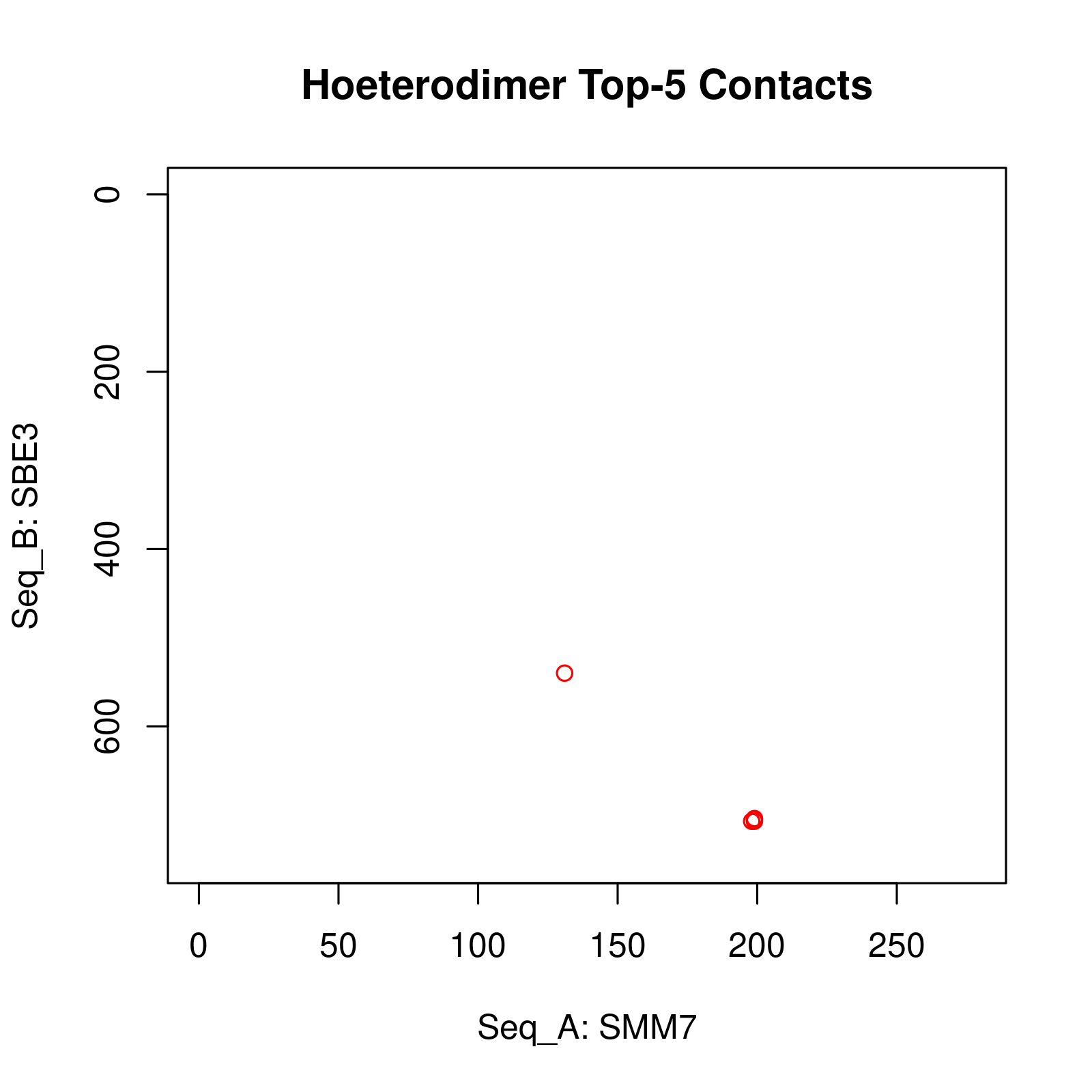
Results of Complex Structure Prediction for Target Name: SMM7_SBE3 ( Click  )
)
Protein sequence Chain A
| 1-60: | M | A | L | A | F | T | S | R | V | A | S | R | P | Q | L | H | A | R | S | A | A | S | H | A | A | A | V | P | A | V | A | R | P | A | T | P | A | T | V | S | A | A | A | P | S | S | S | T | R | S | S | G | V | A | C | R | A | V | R | M |
| 61-119: | D | A | E | L | Q | A | L | L | E | G | A | M | A | R | S | S | C | T | V | G | D | R | V | G | I | I | G | L | K | F | A | E | E | G | Q | K | E | G | C | P | V | M | E | S | A | I | R | L | S | I | Y | L | R | D | Y | K | A | S | D | V |
| 121-179: | R | G | H | S | I | L | E | L | G | T | G | I | G | V | A | G | L | T | L | A | A | F | G | A | H | V | L | L | T | D | L | P | E | M | V | P | V | S | Q | R | N | V | K | K | N | V | D | L | V | R | G | A | G | G | S | A | Q | V | A | A |
| 181-239: | L | D | W | S | S | P | P | Q | E | L | V | E | E | P | W | E | M | I | M | G | S | D | L | V | F | D | Q | A | S | A | E | L | L | A | P | L | L | A | R | L | L | A | G | N | P | G | A | A | V | Y | L | G | H | V | H | R | S | A | A | V |
| 241-278: | D | D | H | M | T | H | V | F | G | Q | H | G | L | A | I | S | A | A | P | R | A | E | G | D | A | H | D | E | D | I | S | I | I | T | V | R | R | A |
Protein sequence Chain B
| 1-60: | M | E | Y | V | D | E | Y | G | P | I | S | P | I | P | N | H | D | G | T | M | C | F | K | W | D | N | T | L | W | S | A | A | E | H | F | K | Y | R | W | N | V | F | K | N | I | R | A | A | I | D | Q | N | E | G | G | F | E | K | F | S |
| 61-119: | Q | G | Y | K | Y | Y | G | F | N | R | G | E | C | N | G | Q | T | G | I | W | Y | R | E | W | A | P | G | A | K | A | L | A | L | V | G | D | F | N | N | W | A | P | A | E | G | H | W | A | F | K | N | Q | Y | G | T | W | E | L | F | L |
| 121-179: | P | D | K | P | D | G | T | S | A | I | P | H | R | T | K | V | K | A | R | I | E | G | A | D | G | S | W | Q | D | K | I | P | A | W | I | K | W | S | T | Q | E | W | N | E | V | L | F | N | G | V | Y | W | D | P | P | E | K | G | A | P |
| 181-239: | G | E | V | D | P | D | K | Q | Y | T | F | K | Y | P | R | P | P | K | P | R | A | L | R | I | Y | E | C | H | V | G | M | S | S | E | E | P | K | V | N | S | Y | L | E | F | R | R | D | V | L | P | R | I | R | A | L | G | Y | N | A | I |
| 241-299: | Q | I | M | A | I | Q | E | H | A | Y | Y | G | S | F | G | Y | H | V | T | N | F | F | G | V | S | S | R | C | G | T | P | E | E | L | K | A | L | V | D | E | A | H | R | M | G | I | I | V | L | M | D | I | V | H | S | H | A | S | K | N |
| 301-359: | T | N | D | G | I | N | M | F | D | G | T | D | G | M | Y | F | H | G | G | P | R | G | N | H | W | M | W | D | S | R | C | F | N | Y | G | N | W | E | T | M | R | F | L | L | S | N | A | R | W | W | M | D | E | Y | K | F | D | G | Y | R |
| 361-419: | F | D | G | V | T | S | M | M | Y | H | H | H | G | L | S | Y | T | F | T | G | N | Y | G | E | Y | F | G | M | N | T | D | V | D | A | V | V | Y | L | M | L | V | N | N | L | L | H | D | L | F | P | N | C | V | T | I | G | E | D | V | S |
| 421-479: | G | M | P | A | F | C | R | P | W | Q | E | G | G | V | G | F | D | Y | R | L | Q | M | A | I | A | D | K | W | I | E | V | M | K | L | H | D | D | Y | A | W | N | M | G | N | L | V | H | T | L | T | N | R | R | Y | A | E | A | C | V | G |
| 481-539: | Y | A | E | S | H | D | Q | A | L | V | G | D | K | T | I | A | F | W | L | M | D | K | D | M | Y | D | F | M | A | V | P | G | H | G | A | Q | S | L | V | V | D | R | G | V | A | L | H | K | M | I | R | L | L | T | I | A | L | G | G | E |
| 541-599: | S | Y | L | N | F | M | G | N | E | F | G | H | P | E | W | I | D | F | P | R | V | D | S | Y | D | P | S | T | G | K | F | V | P | G | N | G | G | S | L | H | L | C | R | R | R | W | D | L | A | D | A | D | F | L | K | Y | K | F | L | N |
| 601-659: | A | F | D | R | A | M | C | H | L | D | K | A | F | G | Y | M | S | A | P | N | T | Y | I | S | R | K | D | E | G | D | K | M | I | V | F | E | R | G | D | L | V | F | V | F | N | F | H | P | G | Q | S | Y | Q | D | Y | R | V | G | C | R |
| 661-719: | E | A | G | P | Y | K | L | V | L | S | S | D | E | E | V | F | G | G | Y | R | N | N | T | K | E | N | D | V | T | F | Q | T | Q | S | G | N | F | D | N | R | P | H | S | F | Q | V | Y | A | P | A | R | T | C | A | V | Y | A | P | A | E |
| 721-747: | W | A | D | K | D | A | D | R | K | P | H | G | I | P | G | L | G | I | K | D | L | G | P | Y | F | S | R |
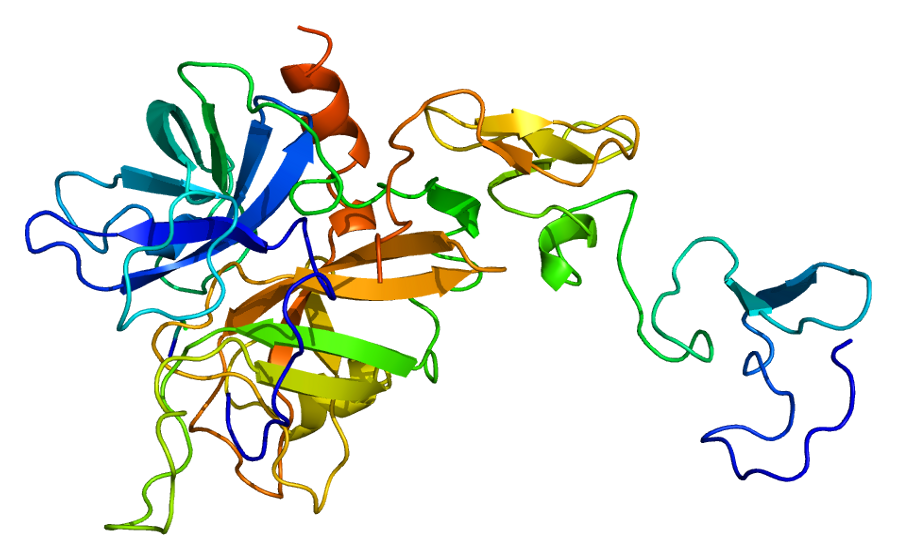
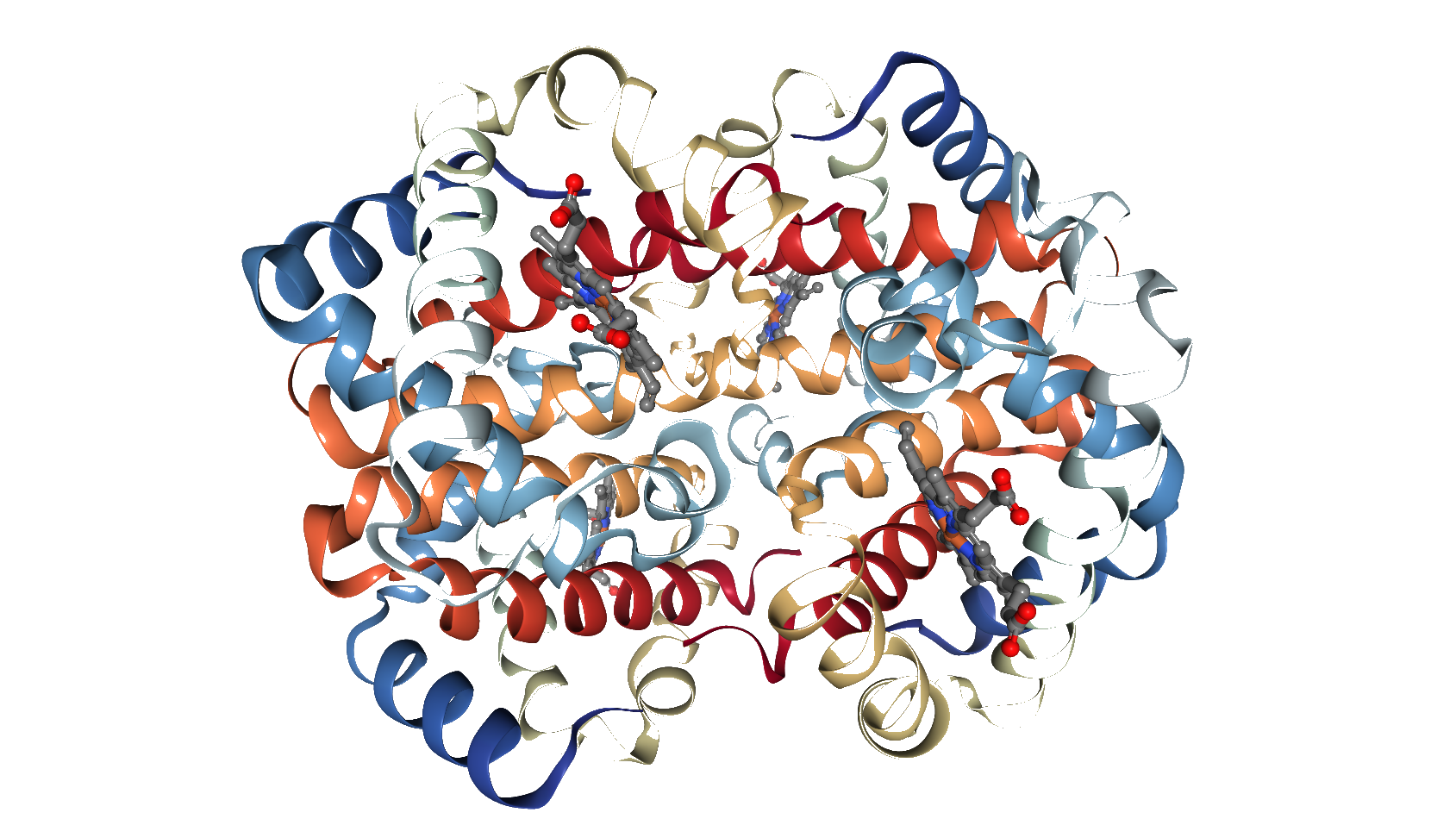
Predicted Top 1 Quaternary structure
|
|
|